| AP Lab 6 Molecular Biology |

Introduction: Exercise 6A: Recombinant DNA Technology
To study the structure and function of a single protein-coding gene, one must prepare the gene in a purified form. Vertebrate cells contain enough DNA to code for more than I 00,000 proteins; therefore it is not very practical to isolate a gene by conventional biochemical procedures. This is why recombinant DNA technology is so important; it can be used to isolate and amplify a specific gene relatively simply.
Plasmids, small circular DNA molecules, are usually extra chromosomal; they exist apart from the chromosomes in most bacterial species. Plasmids are not necessary for the survival of the host bacteria, but they can contain genes that enable the bacteria to survive in certain environments. If a bacterial cell contains a plasmid carrying a gene that confers resistance to antibiotics, then that cell could survive in the presence of the drug.
Plasmids can be introduced into bacterial cells by the process of transformation. Bacteria placed in a calcium chloride solution can take in plasmid DNA molecules. In this way, large amounts of specific plasmid DNA can be prepared, because one transformed cell gives rise to duplicate cells also containing the plasmid DNA molecule. Plasmids are very important for the molecular biologist because they serve as gene- carrier molecules called cloning vectors. A gene of interest can be joined to vector DNA to form a hybrid or recombinant molecule that can replicate in bacteria. When preparing a recombinant DNA molecule, a procedure is required for cutting cloning vectors and cellular DNA molecules in precise positions.
Restriction nucleases are important for recombinant DNA technology because they cut DNA at specific sites. These enzymes are usually made by bacterial species in which they degrade invading foreign DNA within the bacterial cell. Most restriction enzymes recognize a specific sequence of nucleotides in DNA and cut a long DNA double helix into restriction fragments, which are measured in the process of agarose gel electrophoresis.
Introduction: Exercise 6B: DNA Fingerprinting
Electrophoresis is the movement of charged particles in solution under the influence of an electric field. In gel electrophoresis, agarose gel is the stabilizing medium that serves as a matrix for the buffer in which the sample molecules travel. The gel is submerged in buffer within the electrophoretic gel cell. The samples are loaded into the sample wells in the gel, and electric current is passed through the gel.
Molecules of DNA are negatively charged because of negative charges on the phosphate group. In this exercise, nucleic acids migrate through the pores of the gel from the negative end of towards the positive end. The large DNA molecules move more slowly than smaller molecules, therefore molecules are sorted according to size.
Objective: Exercise 6A
Investigate basic genetic concepts by transforming bacterial cells by inserting an ampicillin-resistant gene into E. coli cells.
Objective: Exercise 6B
Investigate basic genetic concepts by using restrictive enzymes to digest phage lambda DNA and separate and identify the DNA fragments using gel electrophoresis.
Materials and Methods: Exercise 6A
The materials used in this exercise included: 2 Luria agar plates, 2 Luria agar plates with ampicillin, 2 microcentrifuge tubes, 1 inoculating loop, 1 Bacti-Spreader, sterile micropipets, calcium chloride, Luria broth, plasmid pUC8, Bunsen burner, hotplate, ice, waterbath.
The two microcentrifuge tubes were marked !I+” and !1-!1, and 250IJI cold calcium chloride was added to each using a pipet. A large colony of bacteria was added to each tube with a sterile inoculating loop. A micropipet was used to transfer 10IJI of the plasmid pUCS solution to the !I+” tube. Both tubes were incubated on ice for 15 minutes, and meanwhile, the two Luria agar plates were labeled “+” and “-” and so were the two Luria plates with ampicillin. The tubes were removed from ice and placed in a 42°C hot waterbath for 90 seconds. The tubes were then removed from the waterbath and placed on ice for two minutes. A micropipet was used to add 250IJI Luria broth to each tube. Another micropipet was used to add 100IJI of the !I+” solution to the two !I+” plates and 1oomicroliters of the solution to the two “-” plates. The bacteria was flamed to sterilize, and after cooling, was used to spread the cells over the entire surface of the plates. After five minutes, the plates were placed in a 37°C incubator, inverted, overnight.
Materials and Methods: Exercise 6B
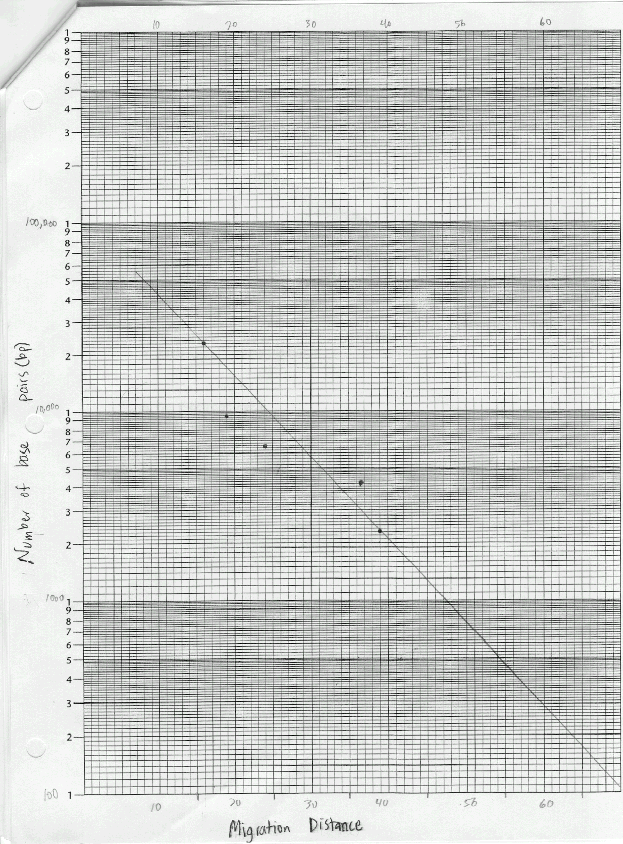
The materials used in this exercise included: 8% agarose gel, 2 electrophoresis chambers, power pack, running buffer- Tris, micropipetter and tips, staining tray, methylene blue dye, gloves, aprons, 4 DNA samples cut with restrictive enzymes, vial tray, microcentrifuge, paper, pencil, distilled water, spatula, plastic container for destaining, masking tape, light box, ruler, semi-Iog graph paper. The gel, on the gel tray, was placed in the center of the chamber, with the well-side of the gel near the black electrode. Approximately 350ml of running buffer were added to the chamber. Of each DNA sample, 10 microliters was loaded into the corresponding gel lane with a micropipet. The power cords were attached to the appropriate connections, and the power supply was turned on, set to 50 volts. The samples were allowed to migrate for three hours. The gel was then removed, stained, and destained overnight. The gel was viewed on a light board, and the band migration distances were measured.
Results: Exercise 6A
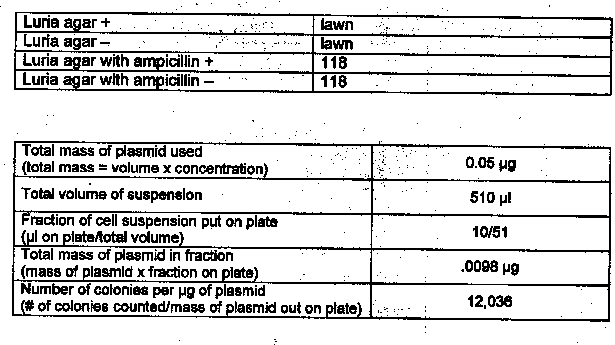
Questions
1. Based on your experimental results, did transformation occur? Why or why not?
Yes, transformation did occur. Colonies of E. coli grew in the presence of ampicillin.
2. What other methods can be used to verify that transformation occurred?
DNA fingerprinting
Results: Exercise 6B

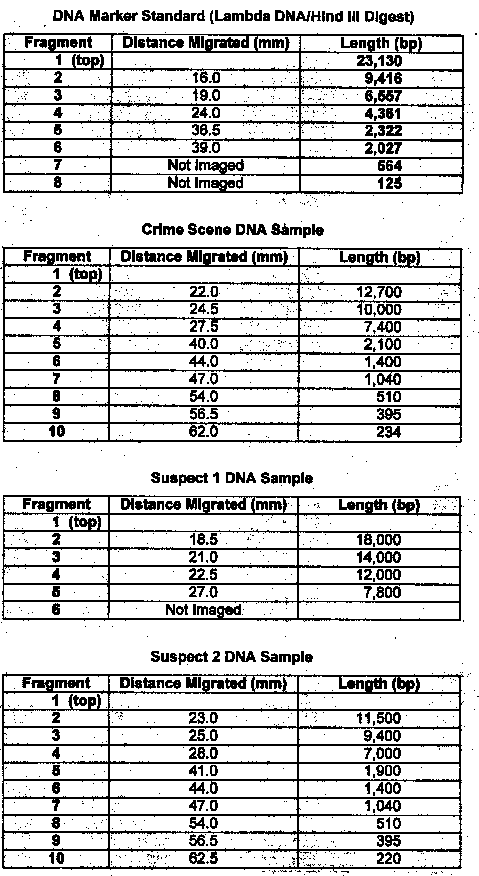
Questions
1. Compare the banding patterns. Do you think the DNA samples were the same?
No, the samples were different sizes
2. Which of the two suspects were the real burglar? Suspect #2
3. Explain the function of each of these steps in DNA fingerprinting:
a. Restriction Enzyme digest – used to cut DNA
b. Gel electrophoresis – used to separate different size pieces of DNA
c. Denaturing into single-stranded DNA – process used to view only one strand of the DNA double helix
d. Southern Blot – DNA bands transferred to a nitrocellulose paper
e. Radioactive DNA probe – used to find & bind to the complementary sequence in one or more RLFP’s
f. Autoradiograph – used to show similarities in DNA samples
Error Analysis
Lab 6A: Not enough agar was poured on the plates.
Lab 6B: Base pair counts for the DNA bands could have been inaccurate as suggested by the best-fit line on the graph.
Conclusion
Lab 6A: This lab showed that genes can be inserted into living bacterial cells thus transforming the cells and giving them new properties such as ampicillin-resistance.
Lab 6B: Through gel electrophoresis of the DNA samples, it was determined that suspect #2 was guilty. Their DNA banding pattern matched the banding pattern taken of the DNA at the crime scene.
| BACK |
